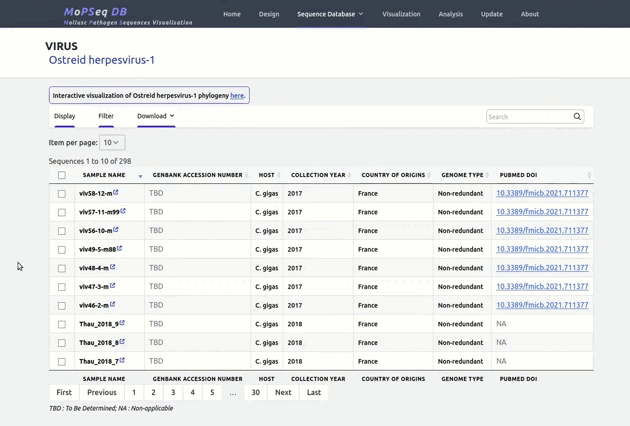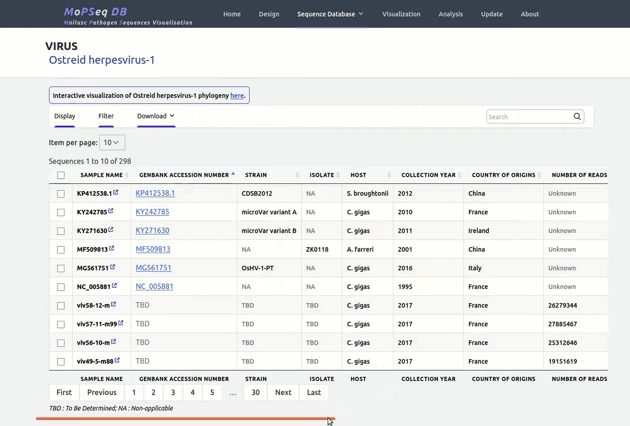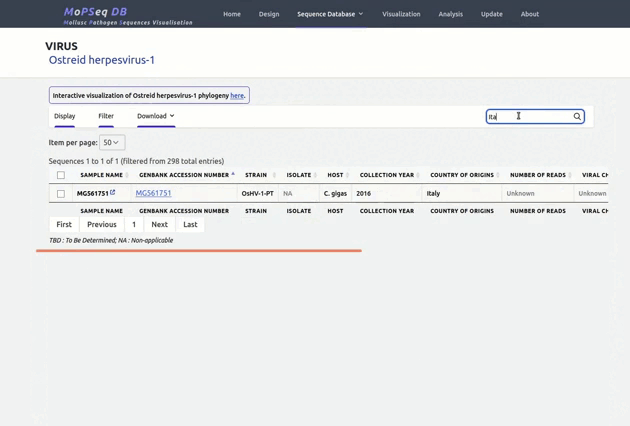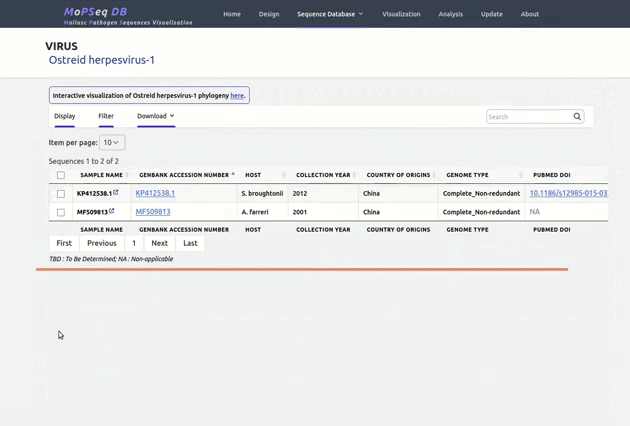
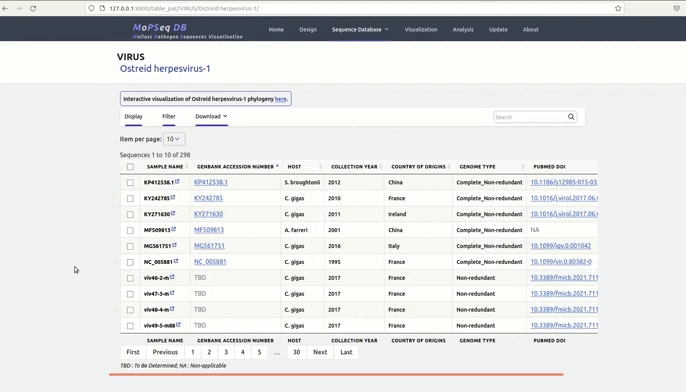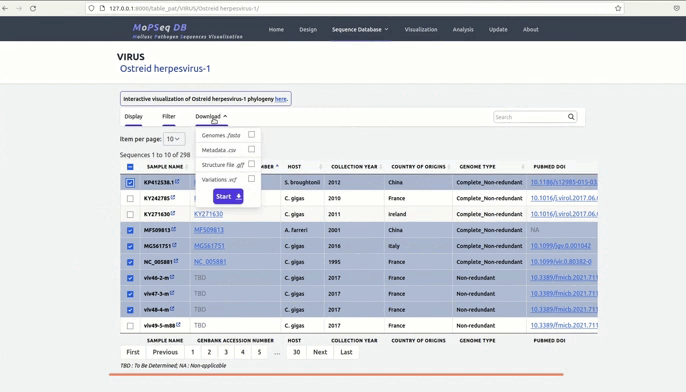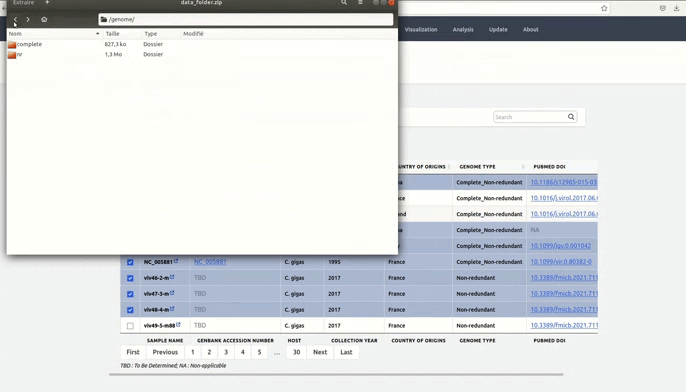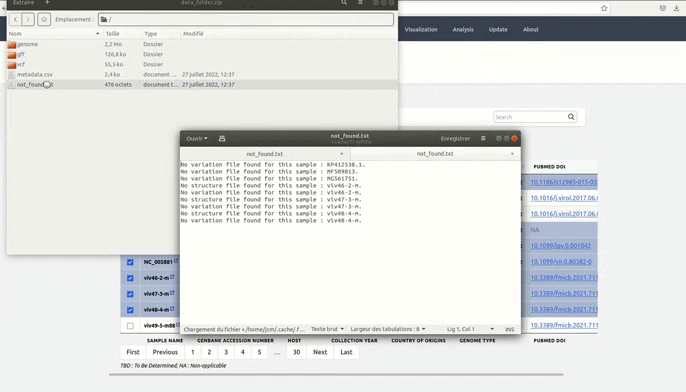
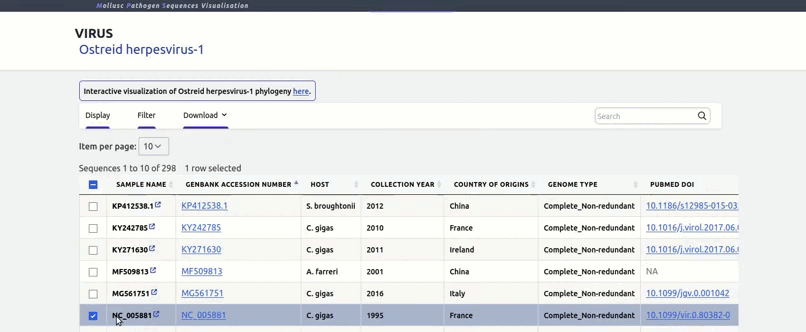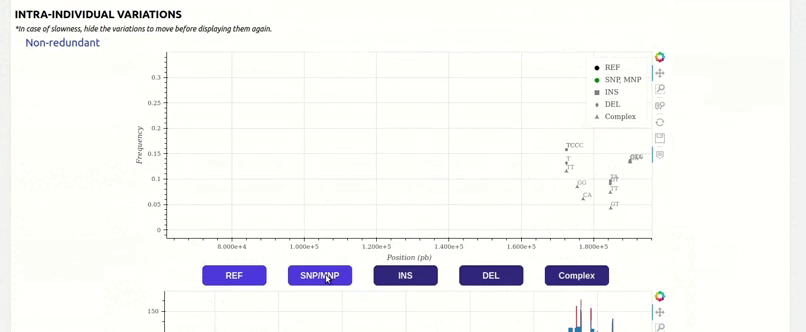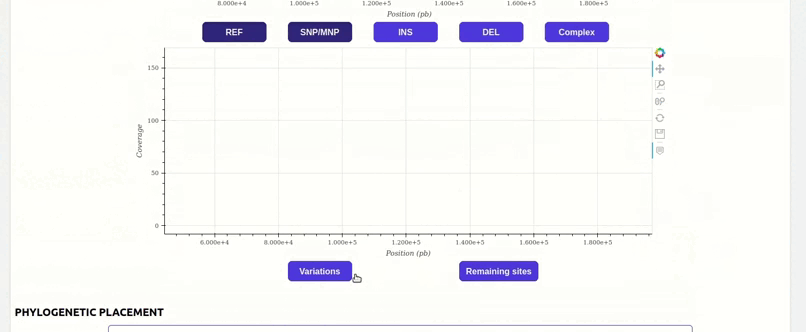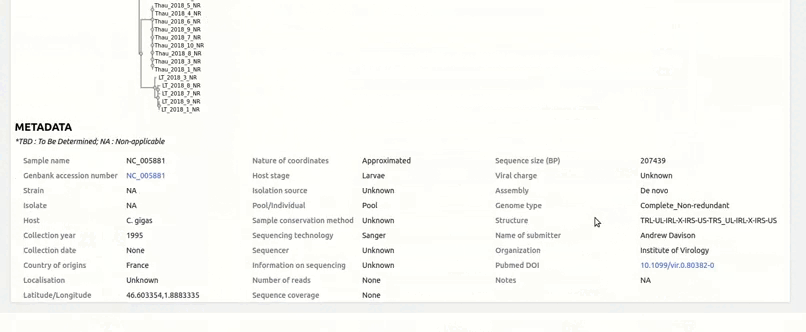
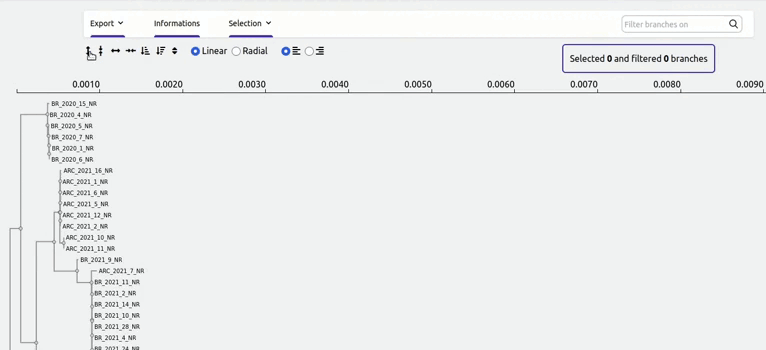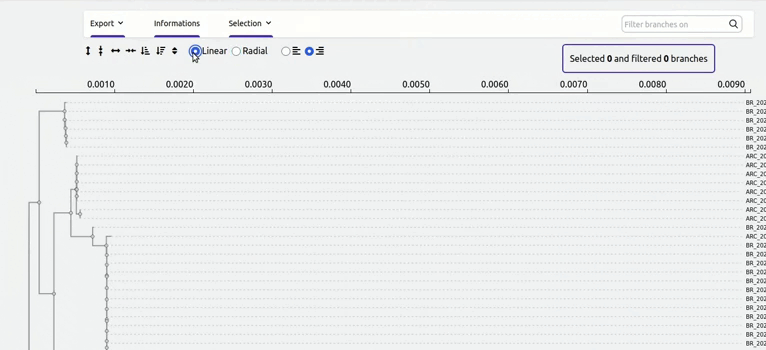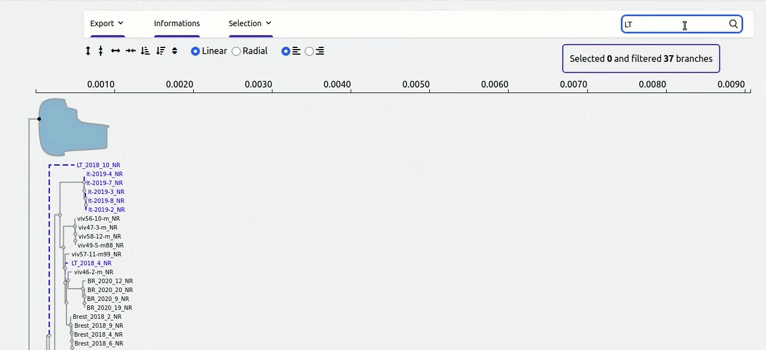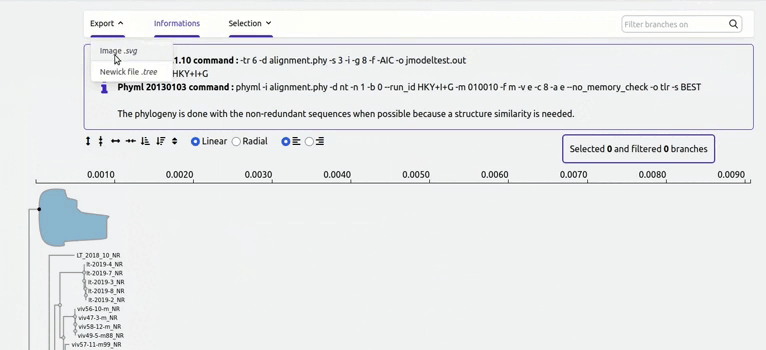
organized metadata, and they are readily visualised.
and metadata files.
Going further:
Upcoming features include support for automatic phylogenetic analysis, specifically the phylogenetic placement of user's sequences. Additionally, the platform will feature an automated submissions tool for new sequences, streamlining the process of adding and updating data.
Furthermore, users will benefit from visualising phylogeographic analyses through the integration of Nextstrain modules (Hadfield et al. Bioinformatics, 2018.), providing valuable insights into the geographic distribution and evolutionary history of pathogens.
These enhancements will further elevate the capabilities of MoPSeq-DB, making it a cutting-edge and comprehensive resource for researchers and practitioners in the field of mollusc pathogen genomics. We look forward to delivering these exciting features to the scientific community and advancing our collective understanding of pathogen surveillance and diagnostics.


